Team Julie PERROY
Pathophysiology of synaptic transmission

Through their ability to form and modify their synaptic connections, our neurons are constantly remodelled by our experiences, which modifies our functional neural networks and our interaction with the environment. Our work aims to understand how learning emerges from plasticity, using a multi-scale approach, from the dynamics of synaptic molecules to the cellular and behavioral events that do or do not trigger plasticity. Understanding this plasticity will not only lead to the discovery of new therapies for neurological diseases and psychiatric disorders, but will also provide answers to fundamental questions about cognition.
We are investigating the molecular and cellular determinants that regulate the formation, function and plasticity of synapses, as well as the signalling of ensembles of neurons responsible for learning behavior. This enables us to approach the question of behavioral disorders, whether they result from genetic or environmental factors, from the angle of molecular, cellular and neuronal network mechanisms.
Neuronal plasticity is a constantly active process: neuronal activity modifies synaptic properties, which in turn change the activity of neuronal networks. We are therefore focusing on the dynamics of synaptic proteins involved in glutamate receptor subcellular trafficking, membrane targetting and signalling. Our previous work show that the different functions of a receptor rely on its ability to engage in specific protein-protein interactions with scaffolding and effector proteins to form a functional unit called a receptosome. These protein-protein interactions are finely regulated, in a highly dynamic way, by interaction with the environment, and therefore need to be studied from a systems biology perspective, combining predictions from theoretical models and cutting-edge experimentation. For example, understanding whether a receptor is activated as a free molecule or as part of different complexes and what its causal role is in each of these conditions are key biological issues in understanding the fundamental logic of plasticity. This knowledge also offers the possibility of exclusively targeting the altered signalling pathway in a given pathology.
Our projects aim to characterise the dynamics of synaptic molecules, the logic of the signalling and plasticity system of a neuron (and ensembles of neurons), and the resulting behavioral learning. We work within a formal theoretical framework (biophysical and cognitive modelling) that links these scales. On the one hand, this approach provides an explanatory framework describing how properties at one level of organisation (e.g. neural networks) emerge from lower levels (interactions between neurons, between molecules); on the other hand, it provides quantitative predictions (what form of plasticity, what molecule involved) to guide our experiments, which then answer specific questions. In order to characterise these different scales of organisation (molecules, neuron, network, behavior), we are developing methods for the real-time study of protein-protein interaction dynamics and neuronal signalling in vitro and in vivo, as well as tools for constraining molecular dynamics at synapse level and understanding their physiological and therapeutic importance. Finally, by taking advantage of cell and mouse models, we are promoting mechanistic translational research, going back and forth from the mouse to the patient. To propose therapeutic targets, we can reproduce a genetic mutation identified in a patient with neurodevelopmental disorders and/or expose the mice to an environmental factor, in order to overcome the correlations between factors and disorders and understand their causality.
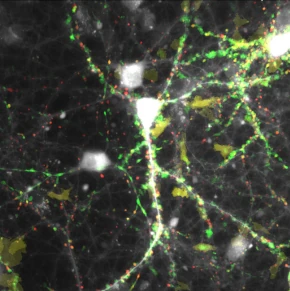
Projection of calcium events recorded over 2 minutes with GCamP6s in hippocampal neurons, sorted according to size and spread

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10-
2011Habilitation to Supervise Research - University of Montpellier - France
-
2001PhD in Cellular Biology, University of Montpellier - France
-
1998MSc in Physiology - University of Montpellier - France
-
2015-Group leader - Institute of Functional Genomics (IGF) - Montpellier - France
-
2020-Research Director at CNRS (DR1) - Institute of Functional Genomics (IGF) - Montpellier - France
-
2012-2020 Research Director at CNRS (DR2) - Institute of Functional Genomics (IGF) - Montpellier - France
-
2004-2012Research Associate at CNRS - Institute of Functional Genomics (IGF) - Montpellier - France
-
2002-2004 Postdoctoral fellow (Dir. M. Bouvier) - Montréal - Canada
-
1998-2001PhD student - CNRS INSERM Center for Pharmacology and Endocrinology (CCIPE) - Montpellier - France
- Awards
-
2015-2021ERC consolidator
-
2002-2004EMBO Fellowship
-
2002Thesis Prize from the French Society for Neuroscience
- Responsabilities
-
2022-2027Expert for Aviesan, ITMO Neurosciences
-
2021-2024Member of the Governing Council of the french society of neurosciences
-
2019-2025Coordinator of the Scientific Committee of the Center of excellence for Autism and neurodevelopmental diseases
-
2017-2022Member of the INSERM Scientific Council
-
2015-2018Member of the ANR CE16 evaluation committee (Mol and Cell Neuroscience)
- The originality of our work lies in the development of innovative tools to elucidate the dynamics and role of protein-protein interactions around receptors. Indeed, we have helped to demonstrate that the various functions of a receptor rely on its ability to interact specifically with scaffolding proteins and effectors to form a functional unit called the receptosome. These protein-protein interactions are finely and dynamically regulated by environmental stimuli.
- My team has been interested in the role of receptosomes for many years, particularly metabotropic glutamate (mGlu) receptors. To elucidate the spatio-temporal dynamics of protein complexes associated with mGlu receptors in neurons, we have developed and used state-of-the-art biophysical approaches. We then constrained the dynamics of protein-protein interactions within the glutamatergic receptosome (using molecular/pharmacological tools or transgenic mice) to understand their functions under physiological and pathological conditions.

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10-
2011PhD - Neurosciences - University of Montpellier - France
-
2008MSc - Biology & Health - University of Montpellier - France
-
2022-Research Associate at CNRS (CRCN) - Institute of Functional Genomics - Montpellier - France
-
2016-2021Postdoctoral fellow - Institute of Functional Genomics - Montpellier - France
-
2012-2015Postdoctoral fellow - University of Genena - Switzerland
-
2008-2012PhD student - Institute of Functional Genomics - Montpellier - France
-
2014Price during "Les grandes avancées du Pôle Rabelais" - Montpellier
- I'm a neuroscientist specialized in the study of the molecular mechanisms that modulate synaptic plasticity. I'm interested in the sequences of events that take place in the dendritic spines of glutamatergic synapses during plasticity, from changes in protein interactions and protein localization to morphological changes of the spines.
- The long-term goal is to better understand the role of molecular dynamics associated with the mGlu5 receptor in memory location, with a particular focus on mouse models of Autism Spectrum Disorder. Indeed, there are models of ASD in which certain proteins associated with the mGlu5 receptor are mutated. These models are real tools for studying the importance of these proteins in plasticity and for developing strategies to modulate their molecular dynamics with the aim of restoring the plasticity defects and certain cognitive disorders associated with these pathologies.
- The techniques I mainly use are:
- Hippocampal primary cultures and slices
- BRET imaging
- Biochemistry
- Molecular biology
- Calcium imaging
- Super-resolution imaging
- Electrophysiology

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10-
2022Habilitation to Supervise Research (HDR) - Sorbonne University - Paris - France
-
2010PhD in Computational Neuroscience - Sorbonne University (ex. Pierre et Marie Curie) - Paris - France
-
2007MSc in Integrative Biology and Physiology (Neuroscience) - Sorbonne University (ex. Pierre et Marie Curie) - Paris - France
-
2021-CNRS Research Fellow - Institute of Functional Genomics - Montpellier - France
-
2016-2021CNRS Research Fellow - Neuroscience Paris Seine - Paris - France
-
2012-2016Postdoctoral fellow - Neuroscience Paris Seine - Paris - France
-
2010-2012Temporary Teaching and Research Associate (ATER) - Institute of Intelligent Systems and Robotics - Paris - France
-
2007-2010PhD Student - Institute of Intelligent Systems and Robotics - Paris - France
-
2023-Elected member of the French Neuroscience Society
-
2022-Elected member of the CNRS National Committee (section 26)
-
2014-2016Elected member of the Laboratory Council - Neuroscience Paris Seine
-
2008-2009Elected student member of the Doctoral School Council - Sorbonne University
-
2007-2009Elected student member of the Biology UFR - Sorbonne University
- I am a neuroscientist with dual expertise in in vivo electrophysiology/behaviour and computer modelling.
- My research interests include:
- neuromodulation, plasticity, linking electrical and behavioural time scales
- multi-level models (intracellular pathways, neuronal biophysics, neuronal networks, cognitive/behavioural)
- the prefrontal cortex and its many partners
- Laboratory techniques
- Level 1 designer and surgery training
- Data analysis
- Sorting and clustering of action potentials (APs)
- Dynamic analyses for discrete (AP) and continuous (EEG, calcium) time series
- EEG/LFP analysis (Fourier, coherence, wavelets, cross-frequency coupling, etc.)
- Behavioural analysis (choices, movements, etc.)
- Computer modelling
- Construction of models
- Fitting models to experimental data (e.g. comparing models)
- Simulations (Matlab, Pyhton)
- Analysis (mean field, bifurcation analysis)

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10-
2011Habilitation to Supervise Research (HDR) - Université Pierre-et-Marie-Curie (UPMC) - Paris - France
-
1994Doctorate in Molecular and Cellular Pharmacology - Paris 6 University - France
-
1991Magistère, École Normale Supérieure de Lyon - France
-
2012-CNRS Research Director (DR2) - Institute of Functional Genomics (IGF) - Montpellier - France
-
1996-2012Researcher at CNRS
-
1994-1996Postdoctoral fellow (Dir. R.Y. Tsien) - Univ. of California San Diego - USA
-
1993Military service (Dir. J.P. Changeux) - Institut Pasteur - Paris - France
-
1991-1994Doctoral student (Dir. A. Marty) - Neurobiology Laboratory - École Normale Supérieure - Paris - France
-
2005-2022Team leader at Sorbonne University
- I am interested in the dynamic aspects of intracellular signaling and the mechanisms of neuromodulatory signal integration in neurons of the central nervous system. In particular, I have analyzed the specificities of the integration of dopaminergic signals via the cAMP/PKA signaling pathway in striatal neurons, with a particular interest in the role played by phosphodiesterases in this integration.
- Biosensor imaging
- Genetically encoded biosensors from the AKAR series, Epac...
- Imaging biosensors in brain slice preparations
- Data analysis
- Development of a software package for the analysis of biosensor imaging data.
- Programming in the Igor Pro environment
- Data management
- Filemaker Pro database for managing and tracking digital laboratory data
- Electrophysiology
- Patch-clamp on primary cells, brain slices, etc.
- Synaptic transmission analysis

 IGF Sud 227
IGF Sud 227
-
2003Engineer in Biochemical and Biomolecular Engineering, Microbial Genetics - INSA (Institut National des Sciences Appliquées) - Toulouse - France
-
2011-CNRS Engineer - Institute of Functional Genomics (IGF) - Montpellier - France
-
2004-2011CNRS Engineer (fixed-term contract) - Center for Research in Cell Biology of Montpellier (CRBM) - France
-
2015-2020Member of the Laboratory Council (appointed) - Institute of Functional Genomics
-
2014-Responsible BSL-1/ BSL-2 labs - Institute of Functional Genomics
- As an engineer, I am responsible for developing and implementing the molecular approaches of the team's projects by combining techniques of biochemistry, molecular and cellular biology to decipher and modulate the signaling pathways of neuronal plasticity and their dysfunctions associated with autism spectrum disorders. In particular, I design and develop a set of molecular tools (biosensors, actuators, TAT peptides) to visualize and manipulate the dynamics of synaptic protein interactions.
- Main techniques mastered
- Molecular biology techniques: cloning (CRISPR, Gibson, Gateway), site-directed mutagenesis, sequencing, PCR, DNA/RNA purification, genotyping
- Protein biochemistry: Production of recombinant proteins, Western-blot, immunoprecipitation, interactions, assay…
- Cell culture: Primary cultures of neurons / cell lines / iPSCs, transfection, transduction
- Lentivirus and AAV production
- Microscopy: Live cell imaging, BRET, Immunolabeling (IF, IHC)
- Use of scientific software (data processing, image acquisition and analysis: Metamorph, AxioVision, ImageJ, statistics: GraphPad Prism)
- Training courses
- Responsible BSL2 / BSL3
- Designer Level 1
- Project Management

 IGF Nord 223
IGF Nord 223 04 34 35 92 10
04 34 35 92 10

 IGF Nord 223
IGF Nord 223 04 34 35 92 77
04 34 35 92 77

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10-
2024Master Bio-Health - Neurosciences - Montpellier University - France
-
2022Animal Physiology and Neurosciences Licence - Montpellier University - France
- PhD student - Institute of Functional Genomics - Montpellier - France
-
2024Master 2 internship (Dir. S.Trouche - Team S.Trouche) - Institute of Functional Genomics - Montpellier - France
-
2023Master 1 internship (Dir. J.Naude - Team J.Perroy) - Institute of Functional Genomics - Montpellier - France
- Complementary Teaching Mission
- Animal behavior (Operant conditioning, Conditionned Place preference, optogenetics..., )
- Immunohistochemistry
- Imaging techniques and analysis (Apotome, QuPath...)
- Model-based analysis (Matlab, Python...)
- Data Analysis (GraphPad, Office...)

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10

 IGF Sud 227
IGF Sud 227 04 34 35 92 10
04 34 35 92 10
ALUMNI
• Sophie Sakkaki – Préparation de l’Agrégation en SVT
• Noémie Cresto – Chercheuse en Neurosciences (Post-doc, IGF Montpellier, France)
• Benoît Girard – Chercheur en Neurosciences (Université de Genève, Suisse)
• Marin Boutonnet – Chercheur en Neurosciences (Post-doc, Humboldt Université de Berlin, Allemagne)
• Roza Szlendak – Chercheuse en Neurosciences (IMC, warsaw, Pologne)
• Federica Giona – Spécialiste recherche et développement (recherche d’emploi, Milan Italie)
• Federica Bertaso – Chercheuse en Neurosciences (CRCN INSERM – IGF Montpellier, France)
• Jeanne Ster – Chercheuse en Neurosciences (CRCN CNRS – IGF Montpellier, France)
• Elise Goyet – Educatrice spécialisée (Lyon, France)
• Fabrice Raynaud – Enseignant chercheur (Université de Montpellier, France)
• Laura Ceolin – Attachée de recherche Clinique (Mediaxe Formation, Paris, France)
• Selma Dadak – Chercheuse en Neurosciences (Sensorion, Montpellier, France)
• Valériane Tassin – Webmaster (France)
• Adeline Burguière – Enseignante cours particuliers (Matha2, Montpellier, France)
• Jonathan Roger – Responsable des opérations cliniques (Paraxel, Montreal, Quebec, Canada)
- Boutonnet, M., Carpena, C., Bouquier, N., Chastagnier, Y., et al. (2024). Voltage tunes mGlu5 receptor function, impacting synaptic transmission. J. of Pharmacol. – in press
- Sakkaki, S., Cresto, N., Chancel, R., Jaulmes, M., Zub, E., Blaquière, M., et al. (2023). Dual-Hit: Glyphosate exposure at NOAEL level negatively impacts birth and glia-behavioural measures in heterozygous shank3 mutants. Environ Int doi: 10.1016/j.envint.2023.108201
- Areias, J., Sola, C., Chastagnier, Y., Pico, J., Bouquier, N., Dadure, C., et al. (2023). Whole-brain characterization of apoptosis after sevoflurane anesthesia reveals neuronal cell death patterns in the mouse neonatal neocortex. Sci Rep doi: 10.1038/s41598-023-41750-w
- Bousseyrol, E., Didienne, S., Takillah, S., Prevost-Solié, C., Come, M., Ahmed Yahia, T., et al. (2023). Dopaminergic and prefrontal dynamics co-determine mouse decisions in a spatial gambling task. Cell Rep doi: 10.1016/j.celrep.2023.112523
- Moutin, E., Sakkaki, S., Compan, V., Bouquier, N., Giona, F., Areias, J., et al. (2021). Restoring glutamate receptosome dynamics at synapses rescues autism-like deficits in Shank3-deficient mice. Mol Psychiatry. doi: 10.1038/s41380-021-01230-x
- Sarazin, M. X. B., Victor, J., Medernach, D., Naudé, J., and Delord, B. (2021). Online Learning and Memory of Neural Trajectory Replays for Prefrontal Persistent and Dynamic Representations in the Irregular Asynchronous State. Front Neural Circuits doi: 10.3389/fncir.2021.648538
- Mota, É., Bompierre, S., Betolngar, D., Castro, L. R. V., & Vincent, P. (2021). Pivotal role of phosphodiesterase 10A in the integration of dopamine signals in mice striatal D1 and D2 medium-sized spiny neurones. British Journal of Pharmacology, 178(24). doi: 10.1111/bph.15664
- Bouquier, N., Moutin, E., Tintignac, L., Reverbel, A., Jublanc, E., Sinnreich, M., et al. (2020). AIMTOR, a BRET biosensor for live imaging, reveals subcellular mTOR signaling and dysfunctions. BMC Biol. Jul 3;18(1.
- Sebastianutto, I., Goyet, E., Andreoli, L., Font-Ingles, J., Moreno-Delgado, D., Bouquier, N., et al. (2020). D1-mGlu5 heteromers mediate noncanonical dopamine signaling in Parkinson’s disease. Journal of Clinical Investigation 130, 1168–1184. doi: 10.1172/JCI126361
- Nair, A. G., Castro, L. R. V., El Khoury, M., Gorgievski, V., Giros, B., et al. (2019). The high efficacy of muscarinic M4 receptor in D1 medium spiny neurons reverses striatal hyperdopaminergia. Neuropharmacology (2019) 1. doi: 10.1016/j.neuropharm.2018.11.029
Dynamics of the mGlu5 receptosome: implications for synaptic transmission and plasticity
Principal investigator
Enora MOUTIN

Cellular and mouse models enable a better understanding of the molecular and cellular mechanisms behind behavioral disorders and to test new therapeutic approaches. Several clinical situations are being studied in the laboratory.
- Autism spectrum disorders (ASD): Neurodevelopmental and/or cognitive disorders are caused by mutations in genes coding for synaptic proteins, such as glutamate receptors (GRIN2B), scaffolding proteins that control their function (SHANK3) or factors regulating gene expression during plasticity (FMR1). We are also studying the impact of environmental factors on brain function, factors as varied as exposure to pesticides (Glyphosate), viruses (SARS-CoV-2) or stress.
- Subarachnoid haemorrhage: Subarachnoid hemorrhage by aneurysm rupture is often followed by delayed cerebral ischemia, notably grieving patients prognosis. Pathophysiology of this complication is currently unclear. We aim at exploring, in an experimental model, the microvascular defects and the microglial response to subarachnoid hemorrhage using in vivo microscopy, MRI, tissue clearing and transcriptomics. In a translational approach, we also study markers of these phenomena in patients using MRI and dynamic PET imaging.
- General anaesthetic agents: General anesthesia triggers drastic changes in neuronal activity throughout the entire brain. This may be accompanied by mid/long-term changes in cellular functions. We aim to study a few signaling pathways of interst using tissue clearing methods and immunohistochemistry. Relevant identified structures are then characterized using a panel of approaches including in vivo extracellular electrophysiological recordings.
Funding
• ANR (2023-2027) LEARN “Light on intracellular signaling Events shAping functional netwoRks in spatial mapping”
• ANR (2023-2027) IsiBRAIN “Impact of SARS-CoV-2 Infection on the central nervous system”
• ANR (2024-2028) TimeTag “Timed decisions: underlying neural mechanisms and TAGging of neuronal ensembles in the prefrontal cortex”
• ANR (2020-2024) Glyflore “Impact of a chronic dietary exposure to low-doses of glyphosate on the gut microbiota and microbiota-associated physiological functions”
• CCA-Inserm-Bettencourt (2022-2026) “Role of microcirculatory damage in delayed cerebral ischaemia complicating aneurysmal subarachnoid haemorrhage”
• FRC (2024-2026) “Multiscale scientific method to fully characterize the functional consequences of GRIN variants, from biological mechanisms to brain diseases”
• FRM (2023-2026) “Role of mGlu5 receptosome dynamics in plasticity, metaplasticity and flexibility – Alleviation of autism spectrum disorders by molecular tinkering”
• MUSE (2020-2024) “Environmental pesticides and genetic predisposition: a path toward autism spectrum disorders “
• MUSE (2024) STRESSA “Combined effects of environmental stress and genetic predisposition to Autism Spectrum Disorders”
Collaborations
 • Cathie Ventalon (IBENS, Paris France)
• Cathie Ventalon (IBENS, Paris France)
• Raphael Gaudin (IRIM, Montpellier France)
• Stéphanie Trouche (IGF, Montpellier France)
• Bruno Delord (ISIR, Paris France)
• Nicola Marchi et Etienne Audinat (IGF, Montpellier France)
• Sandrine Ellero-Simatos et Laurence Payrastre (INRAe, Toulouse France)
• Dorota Hoffman (IMC, Warsaw, Poland)
• Chiara Verpeli et Carlo Sala (CNS, Milano Italie)
• Amaria Baghdadli (CRA, CHU Montpellier France)
• Hélène Hirbec (IGF, Montpellier France)
• Noura Faraj (LIRMM, Montpellier France)
• Christophe Goze-Bac (L2C, Montpellier France)
• Bertrand Wattrisse (LMGC, Montpellier France)
• Emmanuelle Le Bars (I2FH, Montpellier France)
• Denis Mariano-Goulart (CHU, Montpellier France)
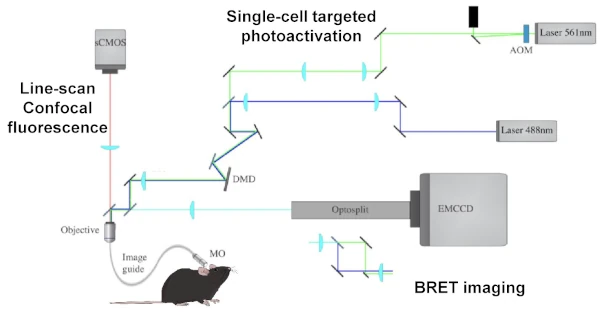
The tools and methods currently available limit our ability to study the dynamics and function of protein-protein interactions and neuronal signalling. To overcome these technological limitations, we are using and developing :
- biosensors for signalling pathways involved in neuronal plasticity (Fluorescence, BRET, FRET), with the ambition of combining readings from several signalling pathways (multiplexing), simultaneously.
- actuators to force, prevent or repair synaptic molecular dynamics and understand their physiological and therapeutic relevance.
- Plugins for automating image acquisition and analysis, to resolve the complexity of a single signal and the intricacies of multiple stimuli.
- new technologies, such as BRET imaging in living cells for subcellular localisation of protein dynamics in real time, as well as in vivo BRET for recording the signalling of neuronal ensembles in animals during behavior.