Team Laurent JOURNOT
Functional genomics of imprinted genes

The Journot Lab is a multi-disciplinary lab that combines molecular biology, (stem) cell biology, genomics, and statistics to shed light on the biological functions of imprinted genes. We focus on 1- the function of the ZAC1/PLAGL1 imprinted transcription factor, 2- the role and the regulation of the imprinted gene network, and 3- the impact of imprinted genes on cortical development. Inspired by the study of imprinted genes, we are developing statistical methods to analyse allelic bias in transcriptomic data.
Parental genomic imprinting is an epigenetic mechanism of gene regulation that restrains the expression of a gene to one allele depending on its parental origin. Genomic imprinting is essential for mammalian development. It targets ~150 genes in placental mammals (eutherians) and less than a dozen in marsupials (metatherians); it does not affect egg-laying mammals (prototherians) and other clades (birds, reptiles, amphibians, fishes…).
The mechanisms that account for the mono-allelic, parent-of-origin-dependent expression of imprinted genes have been studied in details (Hanna & Kelsey, Heredity (Edinb), 2014; Ferguson-Smith, Nat Rev Genet, 2011). A large number of imprinted genes cluster at a limited number of imprinted loci. The repression of one of their parental alleles is controlled by the so-called Imprinting Control Regions (ICRs), which usually comprise a Differentially Methylated Region (DMR). The (de)methylation of the paternal and maternal DMRs governs the expression/repression of the neighboring imprinted genes.
Imprinting defects at defined loci result in different syndromes with complex phenotypes: Silver-Russel, Angelman, Prader-Willi, Beckwith-Wiedemann and TNDM (Transient Neonatal Diabetes Mellitus). In addition, a number of imprinted genes are involved in tumour formation as oncogenes or tumour suppressor genes.
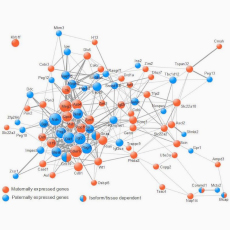
We showed that imprinted genes belong to a network of co-regulated genes we named the imprinted gene network (IGN) (Varrault et al., Dev. Cell, 2006). We next found that the IGN is regulated at the transition from proliferation to quiescence and differentiation during fibroblast cell cycle withdrawal, adipogenesis in vitro, and muscle regeneration in vivo. The IGN also includes bi-allelically expressed genes, notably genes controlling the composition of the extracellular matrix. Our observations suggest that imprinted genes are involved in a common biological process that may account for their seemingly diverse roles in embryonic development, obesity, diabetes, muscle physiology, and neoplasm (Al Adhami et al., Genome Res., 2015).

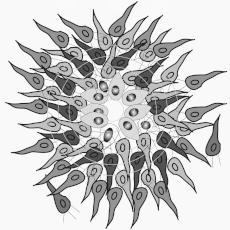
Nestin-positive neural progenitors (green) generated from uniparental mouse embryonic stem cells expressing the cortical marker Pax6 (red) and stained with DAPI (blue).

 IGF Sud 022
IGF Sud 022 04 34 35 92 40
04 34 35 92 40-
1991PhD - Molecular Biology - Université de Montpellier - France
-
1986Master - Biologie Santé - Université de Montpellier - France
-
1985Agrégation de Biochimie-Physiologie (certification for high school professorship)
-
1998-CNRS Research Director - Institute for Functional Genomics - Montpellier - France
-
1992-1998CNRS Staff Scientist - Institute for Functional Genomics - Montpellier - France
-
1991-1992Postdoctorat fellow - Heidelberg University - Heidelberg - Germany
-
1987-1991PhD - Centre CNRS INSERM de Pharmacologie-Endocrinologie - Montpellier - France
- Awards
-
1991EMBO fellow
-
1995CNRS Bronze Medal
-
1997Award from la Ligue Nationale contre le Cancer
-
2000Award from the Schlumberger Foundation for Education and Research
- Scientific appointements
-
2011-2020Director of ''BioCampus Montpellier'' (CNRS-INSERM-Université de Montpellier)
-
2000-2024Director of the MGX-Montpellier GenomiX core facility
-
2007-2026Group Leader: Functional Genomics of Imprinted Genes - IGF Montpellier
-
1998-2007Group Leader: Proliferation-Differentiation-Apotposis - IGF Montpellier
- I am a molecular biologist, I model human diseases in vitro and analyze these models using bioinformatics and statistics.
- My scientific interests and expertise include :
- Parental genomic imprinting.
- The genetic control of proliferation, quiescence, and differentiation.
- Epigenetics, transcriptomics, NGS, gene engineering

 IGF Sud 022
IGF Sud 022 04 34 35 92 40
04 34 35 92 40-
2023HDR - University of Montpellier - France
-
2002PhD - Biologie Santé - University of Montpellier - France
-
2009-Permanent Researcher INSERM - Institut de Génomique Fonctionnelle - Montpellier - France
-
2006-2009Postdoc - Brussels - Belgium
-
2002-2006Postdoc - Bristol - United Kingdown
-
1998-2022PhD student - Centre CNRS INSERM de Pharmacologie-Endocrinologie - Montpellier - France
-
2008-2009Marie Sklodowska-Curie fellow
- I am a molecular biologist, I model human diseases in vitro and analyze these models using bioinformatics and statistics.
- My scientific interests and expertise include :
- Parental genomic imprinting.
- The genetic control of proliferation, quiescence, and differentiation.
- Epigenetics, transcriptomics, NGS, gene engineering

 Fac de Pharma
Fac de Pharma 04 11 75 96 83
04 11 75 96 83

 Fac de Pharma
Fac de Pharma


 IGF Sud 022
IGF Sud 022 04 34 35 92 40
04 34 35 92 40

 Fac de Pharma
Fac de Pharma 04 11 75 96 82
04 11 75 96 82

 IGF Sud 022
IGF Sud 022 04 34 35 92 40
04 34 35 92 40-
2015Engineer in Biology - EICNAM - Montpellier - France
-
2009Bachelor of sciences in Biology - CNAM - Montpellier - France
-
2002Specialization in biomedical research - ESTBA - Paris - France
-
2001Technological University Degree (DUT) in Biological and Biochemical Analyzes - IUT - Montpellier - France
-
1999Baccalaureate: Scientific, specialization in Biology – High school Jeanne d’Arc - Millau - France
-
2024-CNRS Engineer - Institute of Functional Genomics - Montpellier - France
-
2015-2024CNRS Assistant Engineer - Institute of Functional Genomics - Montpellier - France
-
2012-2015CNRS Technician (TCE) - Institute of Functional Genomics - Montpellier - France
-
2002-2012CNRS Technician (TCN) - Institute of Functional Genomics - Montpellier - France
-
2001Lab Technician (2 months CDD) - Hospital - Saint-Affrique - France
-
2025-Prevention staff at IGF
-
2020-2025Member of equipment committee at IGF
-
2010-Head of several common devices at IGF
-
2007-Head of quantitative PCR devices at IGF
- I am an engineer in a research team. My work is organized around two axes:
- Scientific part: conduct a set of specialized techniques in cellular, molecular biology, and genomics as part of the team's projects
- Management part: training in the team and for the common devices for which I am responsible, tutoring BTS students, inventory and order management, database management (Excel), etc.
- Laboratory techniques:
- Cultures of cell lines and primary, human and murine cells
- Molecular biology: PCR, quantitative PCR, Cloning (sub-cloning, Gibson Assembly, CRISPR/Cas9, etc.), Western Blot, etc.
- Cell biology: immunofluorescence, flow cytometry, microscopy, etc.
- Data analysis:
- Specific Software: ZEN, ApE, SnapGene, Flowing Software, Geslab, PrimerBLAST, etc.
- Mastery of Pack office
- GraphPad Prism
- Image J

 IGF Sud 022
IGF Sud 022 04 34 35 92 40
04 34 35 92 40

 IGF Sud 022
IGF Sud 022 04 34 35 92 40
04 34 35 92 40
- Kumar P, Courtes M, Lemmers C, Le Digarcher A, Coku I, Monteil A, Hong C, Varrault A, Liu R, Wang L, Bouschet T. Functional mapping of microRNA promoters with dCas9 fused to transcriptional regulators. Front Genet. 2023 May5;14:1147222. doi: 10.3389/fgene.2023.1147222. PMID: 37214422
- Janbain A, Reynès C, Assaghir Z, Zeineddine H, Sabatier R, Journot L. TopoFun: a machine learning method to improve the functional similarity of gene co-expression modules. NAR Genom Bioinform. 2021 Nov 8;3(4):lqab103. doi: 10.1093/nargab/lqab103. PMID: 34761220
- Reynès C, Kister G, Rohmer M, Bouschet T, Varrault A, Dubois E, Rialle S, Journot L, Sabatier R. ISoLDE: a data-driven statistical method for the inference of allelic imbalance in datasets with reciprocal crosses. 2020 Jan 15;36(2):504-513. doi: 10.1093/bioinformatics/btz564. PMID: 31350542
- Varrault A, Journot L, Bouschet T. Cerebral Cortex Generated from Pluripotent Stem Cells to Model Corticogenesis and Rebuild Cortical Circuits: In Vitro Veritas? Stem Cells Dev. 2019 Mar 15;28(6):361-369. doi: 10.1089/scd.2018.0233. PMID: 30661489.
- Baudement MO, Cournac A, Court F, Seveno M, Parrinello H, Reynes C, Sabatier R, Bouschet T, Yi Z, Sallis S, Tancelin M, Rebouissou C, Cathala G, Lesne A, Mozziconacci J, Journot L, Forné T. High-salt-recovered sequences are associated with the active chromosomal compartment and with large ribonucleoprotein complexes including nuclear bodies. Genome Res. 2018 Nov;28(11):1733-1746. doi: 10.1101/gr.237073.118. PMID: 30287550
- Varrault A, Eckardt S, Girard B, Le Digarcher A, Sassetti I, Meusnier C, Ripoll C, Badalyan A, Bertaso F, McLaughlin KJ, Journot L, Bouschet T. Mouse Parthenogenetic Embryonic Stem Cells with Biparental-Like Expression of Imprinted Genes Generate Cortical-Like Neurons That Integrate into the Injured Adult Cerebral Cortex. Stem Cells. 2018 Feb;36(2):192-205. doi: 10.1002/stem.2721. PMID: 29044892
- Varrault A, Dantec C, Le Digarcher A, Chotard L, Bilanges B, Parrinello H, Dubois E, Rialle S, Severac D, Bouschet T, Journot L. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res. 2017 Oct 13;45(18):10466-10480. doi: 10.1093/nar/gkx672. PMID: 28985358
- Bouschet T, Dubois E, Reynès C, Kota SK, Rialle S, Maupetit-Méhouas S, Pezet M, Le Digarcher A, Nidelet S, Demolombe V, Cavelier P, Meusnier C, Maurizy C, Sabatier R, Feil R, Arnaud P, Journot L, Varrault A. In Vitro Corticogenesis from Embryonic Stem Cells Recapitulates the In Vivo Epigenetic Control of Imprinted Gene Expression. Cereb Cortex. 2017 Mar 1;27(3):2418-2433. doi: 10.1093/cercor/bhw102. PMID: 27095822.
- Al Adhami H, Evano B, Le Digarcher A, Gueydan C, Dubois E, Parrinello H, Dantec C, Bouschet T, Varrault A, Journot L. A systems-level approach to parental genomic imprinting: the imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Res. 2015 Mar;25(3):353-67. doi: 10.1101/gr.175919.114. Epub 2015 Jan 22. PMID: 25614607
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006 Nov;11(5):711-22. doi: 10.1016/j.devcel.2006.09.003. PMID: 17084362.

Functional characterization of the ZAC1/PLAGL1 transcription factor
Principal investigators
Annie VARRAULT et Laurent JOURNOT
Statistical methods for genomic data
Principal investigators
Christelle REYNES et Laurent JOURNOT




