 We originally cloned murine and human ZAC1 as a Zinc finger transcription factor that induced Apoptosis and cell Cycle arrest in different cell types (Spengler et al., EMBO J, 1997; Varrault et al., Proc Natl Acad Sci USA, 1998; Pagotto et al., Endocrinology, 1999; Bilanges et al., Oncogene, 2001). Abdollahi and co-workers independently cloned the rat orthologue they named Lot1 as its expression was Lost On Tranformation of rat epithelial ovary cells (Abdollahi et al., Cancer Res, 1997; Abdollahi et al., Oncogene, 1999). ZAC1/LOT1 was later found to be part of a 3-gene family of zinc finger proteins that includes PLAG1-Pleomorphic adenoma gene 1, PLAGL2 and ZAC1/LOT1/PLAGL1.
We originally cloned murine and human ZAC1 as a Zinc finger transcription factor that induced Apoptosis and cell Cycle arrest in different cell types (Spengler et al., EMBO J, 1997; Varrault et al., Proc Natl Acad Sci USA, 1998; Pagotto et al., Endocrinology, 1999; Bilanges et al., Oncogene, 2001). Abdollahi and co-workers independently cloned the rat orthologue they named Lot1 as its expression was Lost On Tranformation of rat epithelial ovary cells (Abdollahi et al., Cancer Res, 1997; Abdollahi et al., Oncogene, 1999). ZAC1/LOT1 was later found to be part of a 3-gene family of zinc finger proteins that includes PLAG1-Pleomorphic adenoma gene 1, PLAGL2 and ZAC1/LOT1/PLAGL1.
Is ZAC1 the 6q24 tumor suppressor gene ?
Given its functional properties, its silencing during epithelial transformation, and its chromosomal localization on 6q24, a region known to harbor a tumor suppressor gene, ZAC1 was suggested to be the 6q24 tumor suppressor gene. No inactivating mutation has been identified so far, but loss of ZAC1 expression was observed in various neoplasms (e.g. Abdollahi et al., Oncogene, 1997; Varrault et al., Proc Natl Acad Sci USA, 1998; Bilanges et al., Oncogene, 1999; Pagotto et al., Cancer Res, 2000; Abdollahi et al., J Biol Chem, 2003; Kamikihara et al., Int J Cancer, 2005; Basyuk et al., Mol Cancer Res, 2005). We later showed that constitutive inactivation of Zac1 in mice did not result in tumor formation (Varrault et al., Dev Cell, 2006). Hence, 2 decades after its cloning, it is still not clear whether ZAC1 is the 6q24 tumor suppressor gene.
ZAC1 is the TNDM gene
In contrast to loss of ZAC1 function, gain of ZAC1 function through overexpression is clearly linked to a pathological condition. The demonstration came from 2 independent lines of investigation; 1) the discovery that ZAC1 is maternally imprinted (Piras et al., Mol Cell Biol, 2000; Arima et al., Genomics, 2000) , and 2) the hunt for the imprinted TNDM-Transient Neonatal Diabetes Mellitus gene on 6q24 (Kamiya et al., Hum Mol Genet, 2000; Docherty et al., Diabetologia, 2010). In that respect, we elucidated ZAC1 imprinting mechanism by showing that ZAC1 is maternally imprinted, paternally expressed through the constitutive methylation of its maternal promoter (Varrault et al., J Biol Chem, 2001).
Neonatal forms of diabetes, although not a major threat for the general population compared to T1D or T2D, offer the opportunity to uncover developmentally important genes. Neonatal diabetes (ND) is a monogenic diabetes that encompasses 2 distinct conditions, Transient (TNDM) and Permanent (PND) forms; both conditions are rare (Grulich-Henn et al., Diabet Med, 2010; Iafusco et al., Acta DIabetol, 2012).
In PND, insulin secretory failure occurs in the late fetal or early post-natal period and does not go into remission. Genes mutated in PND include KCNJ11 and ABCC8, which encode the sulfonylurea receptor 1 (SUR1) and KIR6.2 subunit, respectively, of the β-cell ATP-sensitive potassium channel. Alternatively, PND patients display mutations of INS, GCK, or PDX1 genes.
In TNDM, patients are younger at the diagnosis of diabetes and have lower initial insulin requirements. They are more likely to have intrauterine growth retardation and less likely to develop ketoacidosis than patients with PND. Considerable overlap occurs between the 2 conditions, so that TNDM cannot be distinguished from PND based on clinical features early on (Busiah et al., Lancet Diabetes Endocrinol, 2013). At least 85% TNDM patients relapse during adolescence and suffer from permanent diabetes (Busiah et al., Lancet Diabetes Endocrinol, 2013). About 25% of TNDM patients display mutations in the KCNJ11 or ABCC8 genes. The majority of TNDM patients (75%) display an imprinting or cytogenetic defect of the 6q24 chromosomal region (Temple & Shield, J Med Genet, 2002). The critical 6q24 TNDM region harbors a maternally imprinted, paternally expressed gene, ZAC1/PLAGL1, which is the 6q24 TNDM candidate gene (Docherty et al., Diabetologia, 2010). 6q24 TNDM patients have two active ZAC1 alleles as the result of 3 possible mechanisms (Figure 1). How ZAC1 double dosage alters pancreatic β-cell development or function is unknown; we plan to work on this question in the near future.
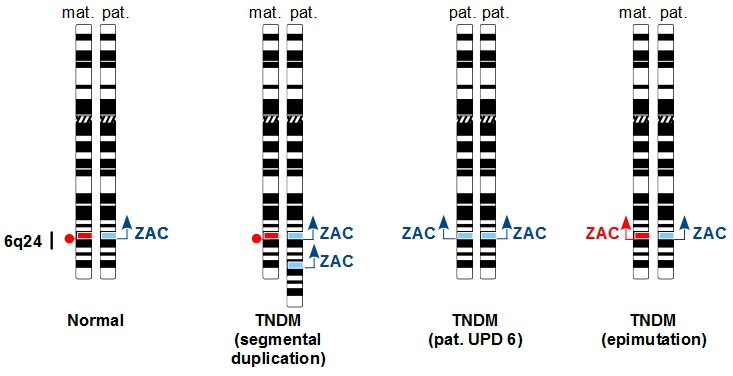
Figure 1. Molecular cytogenetics of 6q24 TNDM. In contrast to normal controls, TNDM patients have 2 active ZAC1 alleles as the result of one of 3 possible mechanisms 1) segmental duplication of paternal 6q24 2) uniparental disomy of the paternal chromosome 6 (pat. UPD 6) 3) hypo-methylation of the maternal 6q24 (epimutation). |
Zac1 target genes
More recently, we aimed at identifying Zac1 target genes genome wide (Varrault et al., Nucleic Acids Res, 2017). We showed that Zac1 up-regulation was associated with physiological cell cycle exit that occurred with contact inhibition, growth factor withdrawal, or cell differentiation. To gain insights into Zac1 mechanism of action, we identified its target genes by combining chromatin immuno-precipitation and genome-wide transcriptomics in transfected cell lines. Zac1-elicited gene regulation correlated with multiple binding to the proximal promoter region through a GC-rich motif. Zac1 target genes included numerous genes involved in signaling, cell adhesion, and extracellular matrix composition, including collagens. Zac1 targets also included 22% of the 409 genes that make up the IGN. Altogether, this work identified Zac1 as a transcription factor that coordinated the regulation of a subset of IGN genes and controlled extracellular matrix composition.
